<< Alignment info. table >>
I : linear weight index for selecting accurate alignments from weak Pareto optimal alignments computed by Cofolga2mo.
P : structure score used in Cofolga2mo.
s : sequence similarity score with affine gap penalties.
<< VARNA usage >>
ZOOM: By rotating the mouse wheel on each VARNA window, user can zoom the structure.
SHIFT: User can drag and shift the structure by cricking the mouse wheel on each VARNA window. If this function does not work well,
please try after ZOOMING the structure.
SAVE and EXPORT: by cricking the left mouse button on the VARNA window, the following menu is displayed.
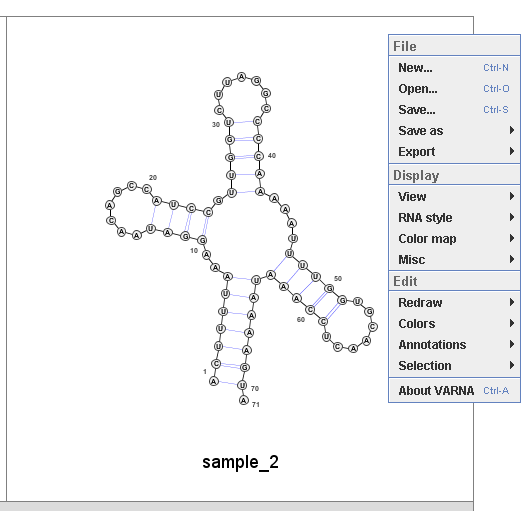
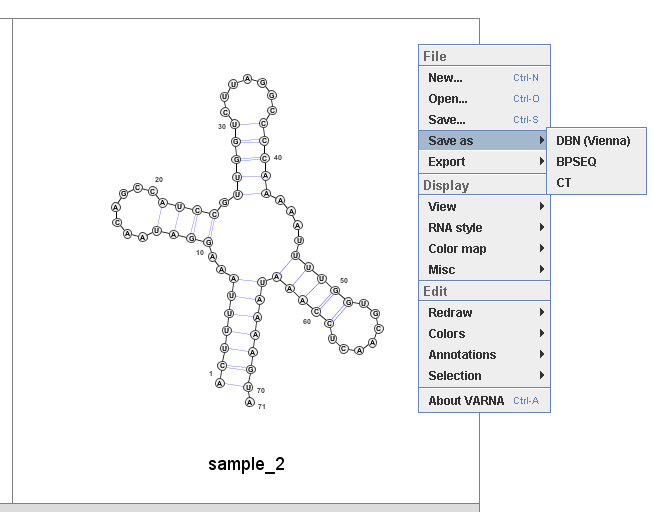
Then user can select 'Save as' to save the structure in a text-based format such as CT (connectivity table format).
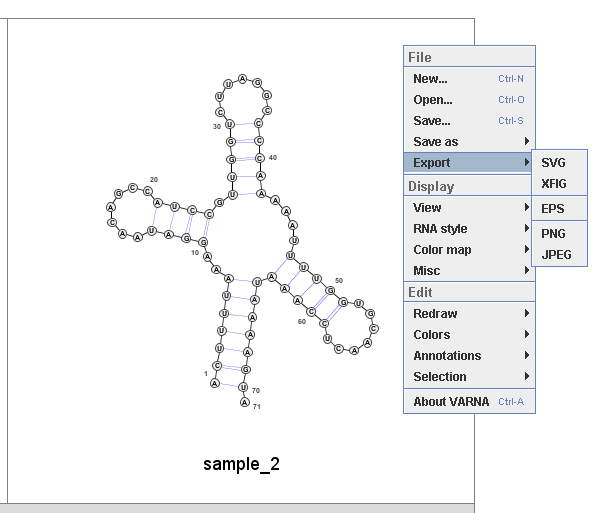
If user selcet 'Export', user can save the structure in SVG, EPS, PNG, and JPEG.
ROTAION: By selecting 'View' and then 'Rotation' in the menu, user can rotate the structure.